Spatial-Live: A Lightweight & Versatile Visualization Tool for Spatial-Omics Data
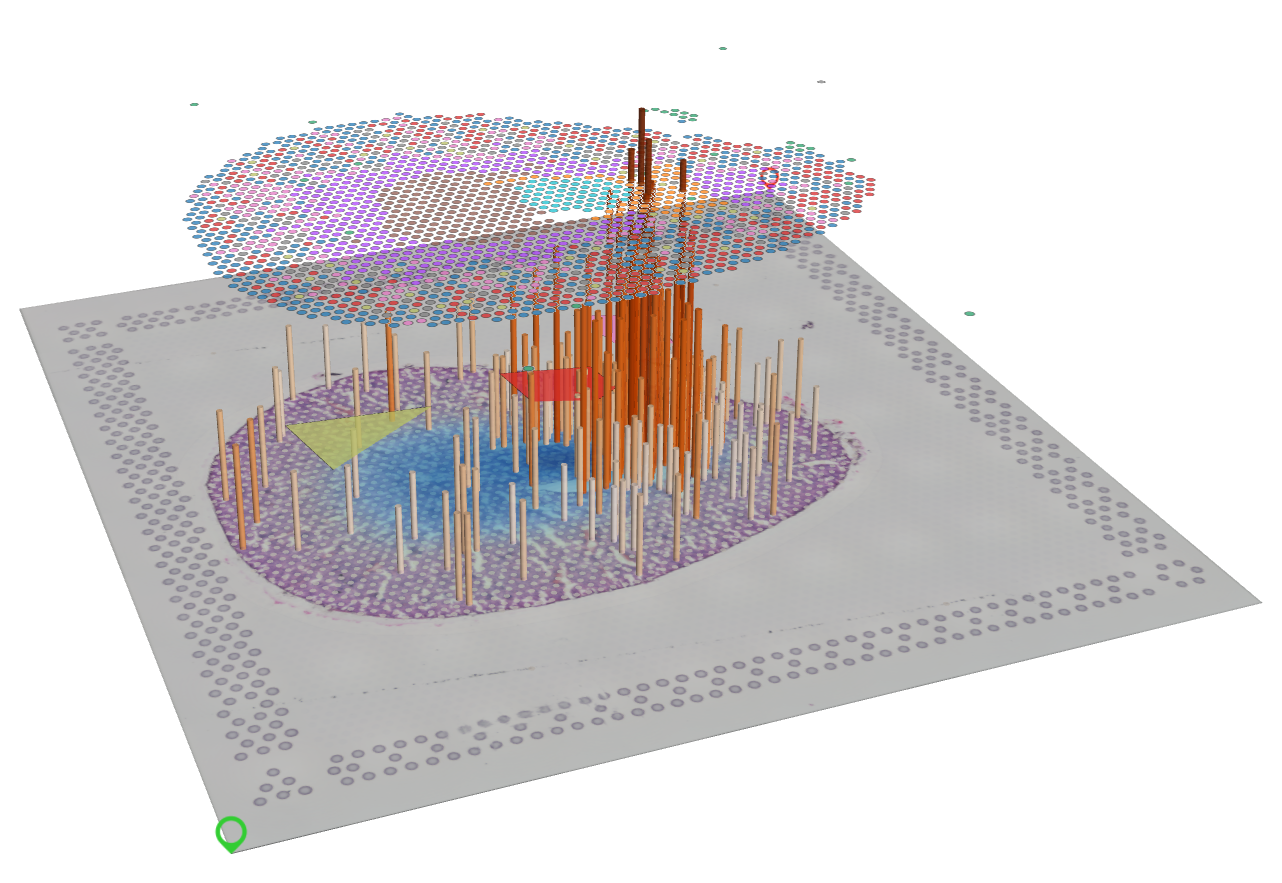
Spatial-Live is a lightweight and versatile visualization tool specifically developed for single cell spatial-omics data analysis, including spatial transcriptiomics and more. Its 3D integration of multiple layers within a single space makes it highly suitable for visualizing multi-type spatial data. Leveraging the rendering capabilities of WebGL2 (GPU) technology, Spatial-Live efficiently processes large datasets, offering interactivity, responsiveness, and a wide range of visualization effects through the stacking of multiple layers. As the saying goes, a picture is worth a thousand words. We firmly believe that effective data visualization plays a pivotal role in data exploration and interpretation, serving as a key component in gaining insights from complex datasets. Spatial-Live represents a valuable tool for achieving these objectives.
The development of Spatial-Live relied on several outstanding third-party libraries, such as deckgl, vue3, and react framework, alongside numerous other open-source libraries that may not all be listed here. I am immensely grateful for the contributions of these libraries and the invaluable work they have provided.
Citation
If you use Spatial-Live in your work, please cite the publication(preprint now) as follows:
Spatial-Live: A lightweight and versatile tool for single cell spatial-omics data visualization
Zhenqing Ye, Zhao Lai, Siyuan Zheng, Yidong Chen.
doi: https://doi.org/10.1101/2023.09.24.559173
Note
This project is under active development.
Contents
- Installation & Usage
- User Guide
- Data Preparation Tutorial - Kidney Visium
- Data Preparation Tutorial - Liver Vizgen
- Python virtual environment setup
- Vizgen MERFISH Mouse Liver data preparation
- Base Image Preprocessing
- Data preprocessing and quality control
- Spatial Distributions of Cells
- Assign Cell Types
- Hepatocyte Zonation: Peri-Portal and Peri-Central zones
- Preparing input files for Spatial-Live
- A screen snapshot from Spatial-Live